Beef Cattle Lymph Nodes Texas am University Prevalence
Introduction
Salmonella enterica subspecies enterica (here after referred to as Salmonella) is an important group of foodborne pathogens resulting in an estimated 1.2 million illnesses, more than 23,000 hospitalizations, and 450 deaths in the United States (US) each year (Scallan et al., 2011). Financial losses resulting from Salmonella infections are considerable; the United States Department of Agriculture (USDA) Economic Research Service (ERS) reports that the total annual cost of foodborne illness attributed to Salmonella is $3,666,600,031 in medical expenses, loss of productivity, and cost of premature death (Economic Research Service (ERS), 2014). Common vehicles of exposure—many of which are presumably attributable to vertical transmission, fecal contamination and poor food hygiene—include eggs, raw milk and dairy products, poultry, produce, and beef (Guo et al., 2011).
Ground beef is an important vehicle for human exposure to foodborne pathogens (including Salmonella), and was implicated in three outbreaks of salmonellosis between 2010 and 2015 (McLaughlin et al., 2006; Centers for Disease Control and Prevention (CDC), 2012b, 2013). Authors have proposed that the carriage of Salmonella by cattle may contribute to the overall prevalence within ground beef products (Bosilevac et al., 2009). It is a common belief that much of the contamination of beef products results from fecal contamination of hides that in turn contaminate the carcass surface during carcass dressing. Consequently, comprehensive food safety systems have been developed and implemented into slaughter processes to mitigate risks associated with surface contamination. In laboratory settings, the same control strategies that have been implemented generally mitigate both E. coli O157:H7 and Salmonella (Wheeler et al., 2014). These interventions appear to have effectively reduced the burden of foodborne illness attributed to E. coli O157:H7 (Centers for Disease Control and Prevention (CDC), 2015); in 2001, the USDA Food Safety Inspection Service (FSIS) recovered E. coli O157:H7 from 0.9% of ground beef samples, whereas in 2014 the recovery was just 0.04%. However, the same successful outcomes have not been observed for Salmonella (Centers for Disease Control and Prevention (CDC), 2015). Despite the implementation of interventions, the prevalence of Salmonella in ground beef products has remained relatively constant and ranges between 1.6 and 4.2% depending on the size of the sample and the analysis methods employed (Bosilevac et al., 2009; Food Safety Inspection Service, 2011).
Because current in-plant intervention strategies primarily targeting surface contamination have not been successful in decreasing the burden of Salmonella—at least to the same extent as E. coli O157, alternative potential routes of contamination other than sanitation carcass dressing procedures have been investigated (Arthur et al., 2008; Haneklaus et al., 2012; Gragg et al., 2013a). Authors proposed that pathogen contamination of ground beef might also occur via incorporation of contaminated lymph nodes, particularly certain peripheral lymph nodes (PLN) (Arthur et al., 2008; Haneklaus et al., 2012; Gragg et al., 2013a; Li et al., 2015). Harborage within the PLN provides Salmonella protection against surface-oriented mitigation approaches, and this source of contamination would explain the greater prevalence of Salmonella observed in ground beef relative to beef trim destined for ground beef. In prior exploratory work, researchers found that recovery of Salmonella from bovine PLN is not uncommon and that likelihood of recovery might be associated with factors such as season (Gragg et al., 2013a). Given the importance of (a) Salmonella as a food-borne pathogen, and (b) the need to target areas of greatest burden, we set out to more fully characterize and describe the burden of Salmonella in PLN in healthy cattle at slaughter by season, region of the country, and production source. This information will be useful in informing risk assessors and targeted risk abatement if and where it is warranted.
Materials and Methods
Subiliac PLN were collected from carcasses of cull and feedlot-fattened (FF) cattle presented for slaughter at commercial abattoirs. The subiliac was selected for it's relatively large size, the convenience with which it can be collected without affecting normal operations in commercial abattoirs, and because it has been in the PLN of choice in most prior studies. Samples were collected over approximately a 1-year period. Samples collected in spring (February through May, 2012) and winter (November and December 2012) were designated as being collected in "cooler seasons," and those sampled in summer/fall (June through October, 2012) were designated as being collected in "warmer seasons." The convenience sample collection included 12 commercial processing abattoirs, including: seven abattoirs that almost exclusively slaughtered cattle fattened in feedlots, three abattoirs that almost exclusively slaughtered cattle removed (or culled) from breeding herds and dairy cattle (i.e., cull cattle), and two abattoirs that slaughtered a mix of FF and cull cattle. Participating abattoirs were categorized regionally (Figure 1) as regions A, B, and C. Three times in each season, a convenience sample of 75 PLN were collected from each of the abattoirs. Following collection, PLN were shipped to either Texas Tech University or the U.S. Meat Animal Research Center for microbiological analyses. All samples were collected from carcasses of animals after post-mortem veterinary inspection at federally inspected commercial abattoirs. As such, the research described herein did not use live animals.
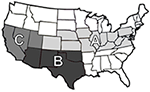
Figure 1. Map of regions based on geographic locations; Region A (Colorado, Illinois, Indiana, Iowa, Kansas, Kentucky, Maryland, Missouri, Nebraska, Ohio, Pennsylvania, Virginia, and West Virginia), Region B (Arizona, New Mexico, Oklahoma, and Texas), and Region C (California, Nevada, and Utah).
Lymph Node Sample Processing and Salmonella Detection
Peripheral lymph nodes were processed as described previously (Brichta-Harhay et al., 2012). Briefly, after the surrounding fat and fascia were trimmed from PLN, the samples were weighed, submerged into boiling water for 3–5 s for surface sterilization, placed in a filtered sample bag (Nasco, Atlanta, GA), pulverized using a rubber mallet, and enriched in 80 mL of tryptic soy broth (TSB; Becton Dickinson, Sparks, MD). Following homogenization, samples were incubated at 25°C for 2 h and then 42°C for 12 h. Enrichments were subjected to immunomagnetic separation (IMS) using paramagnetic beads coated with antibodies to Salmonella (Dynabeads anti-Salmonella, Invitrogen, Oslo, Norway) and the IMS product was transferred to 3 mL of Rappaport-Vassiliadis (RVS; Remel, St. Louis, MO) broth, which was incubated at 42°C for 18 to 20 h. The incubated RVS broth was streaked onto xylose lysine desoxycholate (XLD; Remel, St. Louis, MO) and brilliant green sulfa (BGS; Becton Dickinson, Franklin Lakes, NJ) agar plates prior to incubation at 37°C for 18–20 h.
Quantitative analysis of Salmonella harborage was performed as described previously (Brichta-Harhay et al., 2012; Gragg et al., 2013a), with slight modifications as follows. For this, 1 mL of TSB-PLN homogenate was plated in duplicate to Enterobacteriaceae count plates (EB Petrifilm™; 3M Microbiology, St. Paul, MN; hereafter referred to as EB plates), prior to incubation for enrichment. Enterobacteriaceae count plates were incubated at 37°C for 22 to 26 h; following incubation, colonies were counted. For each EB plate with characteristic gas producing colonies, the plastic film cover with the thin film of agar attached, was removed from the foam backing, and then gently pressed against the surface of an XLD agar plate, essentially replica-plating the colonies present in the EB plate agar. Xylose lysine desoxycholate (XLD) plates were then incubated at 37°C for 16 h and colonies demonstrating typical Salmonella morphology (black colonies with a clear pink ring) were counted and up to 15 colonies per plate were selected for further confirmation. Colonies with morphology atypical of Salmonella on XLD were counted and the number deducted from the original count to arrive at the count used to estimate the concentration of Salmonella present. Peripheral lymph nodes observed to harbor Salmonella after the enrichment step, but at concentrations below the limit of detection of the enumeration methods (~40 colony forming units [CFU]/PLN or 1.6 log10 CFU), were considered to be greater than zero (i.e., Salmonella was present) but at a concentration <1.6 log10 CFU/g of PLN. In these instances a fixed value of 20 CFU/PLN—representing half of the limit of detection—was used as the concentration for data analysis. For each PLN from which Salmonella was recovered, the calculated log10 CFU/PLN was plotted vs. PLN weight (g), in order to examine contamination level as a function of PLN size (as explored by Gragg et al., 2013a), as well as to highlight seasonal and cattle production source trends.
Salmonella isolates were selected from enrichment plates (up to three isolates per PLN) and enumeration plates (up to 15 isolates per PLN) for serotype determination and antimicrobial susceptibility testing. Presumptive Salmonella isolates were confirmed using conventional polymerase chain reaction (PCR) to detect invA (Rahn et al., 1992; Nucera et al., 2006) and molecular serotyping methods (Herrera-Leon et al., 2004). Based on molecular results slide agglutination (O typing) and tube agglutination (flagellar H typing) methods were performed using commercial antisera (Difco, BD Diagnostic Systems, Sparks, MD) to provide a serotype. Where the aforementioned method did not provide resolution, the traditional Kaufmann-White-Le Minor scheme was used for serotyping of isolates (Grimont and Weill, 2007).
Susceptibility to 15 antimicrobial agents was determined using broth micro-dilution (Sensititre CMV2AGNF plates; TREK Diagnostic Systems, Inc., Cleveland, OH) according to manufacturer's guidelines. Isolates were categorized as susceptible or resistant to each antimicrobial based on the breakpoints established by Clinical and Laboratory Standards Institute (2013). All intermediate results were categorized as susceptible for data analysis purposes. Where breakpoints were not available (i.e., ceftiofur and azithromycin), results were interpreted using criteria recommended by Sjölund-Karlsson et al. (2011) or the National Antimicrobial Resistance Monitoring System (Food and Drug Administration (FDA), 2011). Supplementary Table 1 provides a summary of the utilized antimicrobials, abbreviations, breakpoints and antimicrobial classes.
Statistical Analysis
Exploratory data (Gragg et al., 2013a) were used to provide estimates of design prevalence for sample-size calculations. Thirty-five samples per abattoir per time point provided the ability to estimate 30% prevalence within an acceptable margin of error with 95% confidence. Further, 70 samples per plant per season was sufficient to detect Salmonella in one or more PLN with greater than 95% confidence, given a low-end design prevalence of 5%. Assuming β = 0.2 and α = 0.05, the sample size utilized was sufficient to detect relative differences of 50% or greater between seasons, regions, and production sources given a design prevalence of 30% (i.e., in Texas during the summer/fall season) and 5% while allowing for within-abattoir clustering.
A variety of models were constructed to evaluate the association of Salmonella prevalence with season, region, and production source. Generalized linear models were constructed assuming an over dispersed Poisson distribution; an over dispersion parameter was forced into the models to inflate the variance associated with the point estimates. For each sample set, n (the number of PLN from which Salmonella was recovered) was the response variable and the total number of PLN in the sample size (n = ~75) was log-transformed and used as the offset variable. In situations where PLN were harvested from both cull and FF animals in the plant during the same sample collection day, the samples were separated into sets based on the reported production source from which the PLN was harvested during slaughter. Fixed main effects were season, region, and production source. Because of model instability, it was not possible to evaluate the three-way interaction of the main effects. Consequently, and after visual evaluation of the crude means, a main effect of production source was tested and the two way interactions between season and region were evaluated in separate models for cull and FF cattle. Crude means and model estimates were summarized and presented in tabular formats. SAS software (version 9.4, SAS Institute Inc., Cary, NC, USA) was used to perform the statistical analyses. Enumeration data were plotted as total estimated log10 CFU/PLN vs. PLN weight in grams (Figure 2). Data plots were constructed using Prism 5.0d, GraphPad Software, Inc. (www.graphpad.com, San Diego, CA).
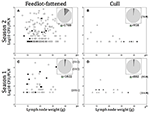
Figure 2. (A–D) Distribution of Salmonella contaminated peripheral lymph nodes (PLN) plotted as Salmonella contamination level (log10 CFU/PLN) vs. PLN weight in grams (g), by season and production source. Numbers in parentheses indicate PLN weight (g) for those found ≥50 g. Pie charts in each (A–D) represent the total number of PLN tested in each season/production source, with light gray representing the proportion tested but negative, medium gray the proportion found positive but not enumerable for Salmonella, and dark gray, the proportion enumerable (>1.6 log10 CFU/PLN). Filled in symbols (O, Δ) indicate particular serotypes observed and include: black, S. Dublin; medium gray, S. Newport; light gray, S. Typhimurium; empty, other serotypes.
Results
Salmonella Prevalence and Enumeration
A total of 5,450 subiliac PLN were collected from cull and FF cattle presented for harvest in three regions of the US, across 11 months. Table 1 provides a summary of the number of sample sets, total number of PLN tested by season, region, and production source the number of Salmonella positive PLN, as well as the crude Salmonella prevalence of PLN collected by season, region, and production source. The overall crude prevalence of Salmonella in PLN from cull and FF animals was 1.8% (33 of 1,840) and 7.1% (256 of 3,610), respectively. The overall crude prevalence of Salmonella in PLN from the cooler season and warmer season were 2.4% (64 of 2,704) and 8.2% (225 of 2,746), respectively. Salmonella prevalence in PLN from cull animals was generally low in every region throughout 11-months (cooler season, 1.7%; warmer season, 1.9%) while that in FF cattle PLN was found to be low in the cooler season (2.7%), yet peaked during the warmer season (11.6%). Abattoirs sampled from Region B that slaughtered cull animals demonstrated a similar prevalence of Salmonella during both seasons (cooler season, 2.1% [adjusted 95% confidence interval 0.64–7.20%]; warmer season, 1.1% [adjusted 95% confidence interval 0.19–5.88%]). PLN collected from FF cattle originating from Region B abattoirs had a prevalence of Salmonella in the cooler and warmer seasons of 6.5% (adjusted 95% confidence interval 3.24–13.16%) and 31.1% (adjusted 95% confidence interval 22.64–42.59%), respectively.

Table 1. Salmonella percent prevalence in subiliac peripheral lymph nodes (PLN) of feedlot-fattened (FF) and cull cattle at harvest by region and season.
In all, Salmonella was recovered from 289 (5.3%) of the 5,450 PLN collected. Of the PLN from which we recovered Salmonella, we were able to quantify the Salmonella concentration in 55.4% (n = 160, or 2.9% of all PLN tested). As demonstrated in Figures 2A–D, the enumerable concentrations ranged from 1.6 (the limit of quantification) to 4.9 log10 CFU/PLN, with 17.6% (n = 51) of positive PLN harboring Salmonella at concentrations >3.0 log10 CFU/PLN. Closer examination of the distribution of PLN harboring higher concentrations of Salmonella revealed that FF cattle in the warmer season (Season 2) were the major contributor to this category, representing 76.5% of enumerable PLN containing >3.0 log10 CFU/PLN. These PLN were predominantly found contaminated with pansuseptible S. Montevideo and S. Anatum and were isolated from FF cattle at harvest in Region B (Figure 2C). Two additional noteworthy observations were: (1) PLN harboring serotypes Typhimurium or Newport were predominantly from cull cattle (Table 2), and that Salmonella concentrations in these PLN were generally low (i.e., ≤1.6 log10 CFU/PLN; Figures 2B,D); and (2) PLN contaminated with S. Dublin were frequently at concentrations >2.0 log10 CFU/PLN and were predominantly contributed by FF cattle at harvest in Region C (Figures 2A,C and Table 2).
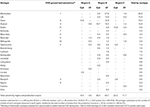
Table 2. Prevalence of Salmonella serotypes isolated from subiliac peripheral lymph nodes (PLN) of cull and feedlot-fattened (FF) cattle at harvest.
Salmonella Serotypes and Antimicrobial Susceptibility Phenotypes
This study resulted in 376 unique Salmonella isolates from 289 PLN found positive for Salmonella. Isolation of multiple unique Salmonella serotypes from a single PLN occurred with 56 samples—4 of cull animal origin and 52 of FF animal origin, with most of the latter originating from the same abattoir in Region B. Occasions where 2, 3, or 4 Salmonella serotypes were isolated from an individual PLN occurred 45, 10, and 1 time(s) respectively. In all 22 serotypes were identified (Table 2); the majority (74.5%) of which were serotypes Montevideo (26.9%), Lille (14.9%), Cerro (13%), Anatum (12.8%), and Dublin (6.9%).
Antimicrobial susceptibility phenotyping identified most isolates, 80.6% (303 of 376) as susceptible to all antimicrobial agents tested. As summarized in Table 3, isolates resistant to two or more antimicrobial classes were infrequently observed (10.7%, n = 40). Tetracycline, sulfisoxazole, streptomycin, and chloramphenicol represented the most common antimicrobials to which Salmonella demonstrated resistance. Salmonella isolates were less frequently resistant to gentamicin, ciprofloxacin, nalidixic acid, and sulfamethoxazole-trimethoprim. Of the 22 serotypes observed in this study, Dublin was the serotype most frequently found resistant to one or more antimicrobials (24 of 26 isolates) and had the greatest diversity of resistance phenotypes (Table 3). Conversely, Salmonella serotypes Cerro, Lille, Anatum, and Montevideo predominantly demonstrated limited resistance phenotypes and were generally pan-susceptible.
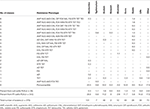
Table 3. Percent of antimicrobial resistance phenotypes among the Salmonella serotypes isolated from subiliac peripheral lymph nodes (PLN) of cull and feedlot-fattened (FF) cattle at harvest.
Discussion
Previous exploratory studies established that cattle PLN can harbor Salmonella (Arthur et al., 2008; Haneklaus et al., 2012; Gragg et al., 2013a). As such, Salmonella may have the potential to circumvent in-plant carcass surface interventions and ultimately represents a human public health burden through PLN inclusion in ground beef product (Koohmaraie et al., 2012; Li et al., 2015). In the study reported here, we were able to more fully characterize and describe the burden across multiple variables. In particular, it is clear that the highest burden (and possible risk) associated with Salmonella harborage in PLN appears to be limited to particular seasons of the year, regions of the country, and cattle production sources. These data show the prevalence of Salmonella in PLN from FF cattle from Region B during the warmer season (31.1%) is responsible for driving the prevalence statistics for FF cattle higher. In fact, FF cattle PLN sampled in Regions A (1.1%) and C (4.8%) remained relatively low with values similar to those observed for cull cattle PLN. Moreover, examination of the serotypes isolated from PLN of FF cattle, especially from Region B, in comparison with serotypes most commonly isolated from ground beef (Table 2), illustrates the degree to which bovine PLN are likely a source of ground beef contaminated by Salmonella. With Salmonella concentrations ranging from 3.0 to 4.9 log10 CFU/PLN in 17.6% of positive PLN in this study—and the possibility of Salmonella harborage in multiple PLN in a given carcass (Gragg et al., 2013b)—the potential for this source of Salmonella to contaminate ground beef is substantial (Li et al., 2015).
An important consideration in the assessment of risk for human illness with regard to Salmonella infection is serotype and antimicrobial resistance. Some Salmonella serotypes are more likely than others to cause severe infection in humans (Jones et al., 2008; Suez et al., 2013). A diverse set of serotypes was isolated in this study; however, the dominant serotypes identified are either infrequently implicated in laboratory-confirmed cases of human salmonellosis (as in the case of Anatum, Lille, and Cerro), or when they are (as in the case of Montevideo) the sources identified are often produce, spices, cheese, or poultry products, as opposed to beef. Despite their documented presence in ground beef, serotypes such as Montevideo and Anatum may pose less of a risk to human health when present in this commodity than serotypes Typhimurium and Newport. Salmonella serotypes Typhimurium and Newport are observed less frequently in bovine PLN surveys, as reported here and in previous studies (Gragg et al., 2013a,b). However, the increased risk of the presence of these serotypes in ground beef resulting in human illness is evidenced by a number of ground beef related outbreaks attributed to serotypes Typhimurium and Newport (Schneider et al., 2011; Centers for Disease Control and Prevention (CDC), 2012b, 2013). In addition, FoodNet data implicate S. Typhimurium and S. Newport as two of the three serotypes responsible for the majority of laboratory confirmed human salmonellosis cases across 10 sites in the US (Centers for Disease Control and Prevention (CDC), 2012a).
When human salmonellosis does occur, it is often self-resolving and only requires care to prevent dehydration. Nevertheless, in approximately 5% of cases—mostly in immune-compromised individuals—life threatening extra-gastrointestinal infections by Salmonella may occur and antimicrobial treatment is then required (Acheson and Hohmann, 2001). Recommended treatment options for salmonellosis include beta-lactams (i.e., penicillin or third generation cephalosporins) or fluoroquinolones (not recommended for children)—and alternative treatment recommendations include sulfamethoxazole-trimethoprim or azithromycin. A majority of our unique strains were pansusceptible; however, 14.3% were resistant to at least one of the antimicrobial classes recommended for treatment of life threatening salmonellosis. Also worth noting, 1.7% of the isolates were resistant to both beta-lactams and fluoroquinolones—which is certainly of concern.
In keeping with this line of thought, the observation here that S. Dublin was detected at enumerable concentrations in PLN of both FF and cull cattle at harvest in Region C (Figure 2 and Table 2)—and was often multi-drug resistant—was unexpected and warrants further investigation. Salmonella Dublin is a cattle-adapted serotype that is commonly detected in ground beef testing programs (Doerscher et al., 2015; Food Safety and Inspection Service (FSIS), 2015). A recent report brought to light the incidence rate of reported S. Dublin infections in humans has been steadily rising in the US since the 1960s, and that these are often cases of bloodstream infections and hospitalization (Harvey et al., 2017). These observations highlight the need for an increased understanding of genes that contribute to Salmonella's success as a pathogen in the human host.
Caution should be taken in interpretation of seasonal-, regional-, and production source-differences observed in the data presented herein, particularly with respect to inferring causal relationships. It is possible that a seasonal association might have been a causal relationship. Alternatively, the association might have been confounded by an unmeasured variable, such as an atypical regional weather event in, for example, Region B. Production source also reflects many unmeasured variables—for example, age of animal at slaughter; in the US, a vast majority of FF cattle are slaughtered at less than 2 years of age, whereas animals culled from breeding herds are typically older with a much wider variation in age. Further complicating the inference on causal relationships is that, in general, animals can only be sampled once, as PLN collection occurs post mortem except in controlled experimental situations. Despite these limitations, consideration of data reported elsewhere infers there are indeed likely causal associations with season, region, and production sources. Gragg et al. (2013a) reported similar associations of Salmonella burden in PLN to those observed here, and Salmonella was recovered from a substantial proportion of PLN collected from FF cattle presented for slaughter in Texas or Veracruz, Mexico (Sofos et al., 1999; Haneklaus et al., 2012; Gragg et al., 2013b; Brown et al., 2015; Cernicchiaro et al., 2016). Moreover, Salmonella is routinely recovered from the feces of cattle in the southern portion of the US (Kunze et al., 2008; Rodriguez-Rivera et al., 2016), yet remarkably, recovery of Salmonella from cattle feces decreases to the north (Wells et al., 2001; Sorensen et al., 2002; Rao et al., 2010; Morley et al., 2011). While these other reports do not preclude confounding of season, region, or production source in the data reported herein, taken together, they do add support that there are indeed predictable differences in Salmonella burden across regions and seasons, at least within North America.
The Salmonella distribution reported herein can aid in identifying production sources that are more likely contributors of Salmonella that may be more important to human health. Targeted removal of large PLN (i.e., the subiliac, superficial cervical, and popliteal) from cattle at harvest is a possible candidate for control; however, removing PLN in the slaughter process is a difficult and imprecise task. Given the limitations of PLN removal in the dressing process, as well as the sporadic nature of PLN harborage, the need for a more precise and efficient mitigation scheme is evident, and yet such methods remain to be defined. Post-harvest strategies, such as irradiation, could decrease Salmonella prevalence in ground beef products, though many consumers have not yet accepted the concept of using methods such as these. Therefore, pre-harvest interventions may be a more practical. Notably, preliminary research testing of a commercially available Salmonella vaccine suggests this form of intervention may decrease the duration of infection in PLN for specific serotypes, in this case Newport (Edrington et al., 2013b). Further research is needed to evaluate the efficacy of these interventions for decreasing the incidence or duration of infection in bovine PLN.
Ultimately, the key to understanding the variation observed in Salmonella harborage in bovine PLN may lay in defining the implications of production source differences in pre-harvest practices. It is unclear by which route Salmonella enters the animal and then is captured within the PLN. One possible explanation is that it escapes the gastrointestinal tract and mesenteric lymph nodes, and then disseminates systemically. Hanson et al. (2016) reported evidence for vertical transmission from the dam to her fetus. Further, serotypes recovered from PLN can vary within an animal, but more closely resembles serotypes recovered from the hides of animals, whereas serotypes recovered from mesenteric lymph nodes more closely resemble those recovered from the feces (Gragg et al., 2013b). It is conceivable, therefore, that a transdermal route of infection could result in accumulation of Salmonella within PLN that receive lymph from the integument. Abrasions or external parasites such as biting insects might, therefore, contribute to the observed burden of Salmonella in PLN (Edrington et al., 2013a,b; Olafson et al., 2016). It is also possible that all of these proposed routes of infection contribute to the burden of Salmonella in PLN; the data presented here illustrate the differences observed in serotype, prevalence, and concentration among FF and cull cattle; these data make it tempting to suggest that harborage of Salmonella in PLN of cull cattle may be a "remnant" of a systemic infection from which the animal recovered—as evidenced by low Salmonella concentrations in PLN with serotypes Typhimurium and Newport. Further, that observed with FF cattle may be more so the result of recent transdermal infection via abrasions or external parasites. Factors such as management (i.e., pest control, animal health best practices, etc.), age of animal at slaughter, and nutrition are all possible drivers of the illustrated differences. Knowledge gaps in the implications of pre-harvest practices ought to be addressed, as they will be instrumental in developing effective and practical solutions to mitigate the food safety risks associated with harborage of Salmonella in bovine lymph nodes.
Author Contributions
Conceived and designed the experiments: GL, TE, and DB-H. Performed the experiments: HW, TB, DB-H, TA, JB, NK, JS, RW, SS, and TW. Analyzed the data and wrote the paper: HW, DB-H, and GL. Critical revision of the paper for important intellectual content: all authors.
Funding
This research was funded by the Beef Checkoff and USDA National Integrated Food Safety Initiative 322 (NIFSI; grant 2011-51110-31081) and USDA Cooperative Agreement (58-6202-2-229).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Names are necessary to factually report available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable. The USDA is an equal opportunity provider and employer.
The authors would like to express their sincere appreciation to the cooperating abattoirs. In addition, we would like to thank Sara Gragg, D. J. Kunze, Kim Kucera, Julie Dyer, Frank Reno, Bruce Jasch, and Greg Smith for their invaluable technical support and Jody Gallagher for her excellent administrative support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02214/full#supplementary-material
References
Arthur, T. M., Brichta-Harhay, D. M., Bosilevac, J. M., Guerini, M. N., Kalchayanand, N., Wells, J. E., et al. (2008). Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J. Food Protect. 71, 1685–1688. doi: 10.4315/0362-028X-71.8.1685
PubMed Abstract | CrossRef Full Text | Google Scholar
Bosilevac, J. M., Guerini, M. N., Kalchayanand, N., and Koohmaraie, M. (2009). Prevalence and characterization of Salmonellae in commercial ground beef in the United States. Appl. Environ. Microbiol. 75, 1892–1900. doi: 10.1128/AEM.02530-08
PubMed Abstract | CrossRef Full Text | Google Scholar
Brichta-Harhay, D. M., Arthur, T. M., Bosilevac, J. M., Kalchayanand, N., Schmidt, J. W., Wang, R., et al. (2012). Microbiological analysis of bovine lymph nodes for the detection of Salmonella enterica. J. Food Protect. 75, 854–858. doi: 10.4315/0362-028X.JFP-11-434
PubMed Abstract | CrossRef Full Text | Google Scholar
Brown, T., Edrington, T., Loneragan, G., Hanson, D., Malin, K., Ison, J., et al. (2015). Investigation into possible differences in Salmonella prevalence in the peripheral lymph nodes of cattle derived from distinct production systems and of different breed types. J. Food Protect. 78, 2081–2084. doi: 10.4315/0362-028X.JFP-15-198
PubMed Abstract | CrossRef Full Text | Google Scholar
Centers for Disease Control and Prevention (CDC) (2013). Multistate Outbreak of Salmonella Typhimurium Infections Linked to Ground Beef [Online]. Available online at: http://www.cdc.gov/salmonella/typhimurium-01-13/ (Accessed August 28 2013).
Centers for Disease Control and Prevention (CDC) (2012a). Foodborne Diseases Active Surveillance Network (FoodNet), Table 5 FoodNet–Number and INCIDENCE of Salmonella Infections by Serotype 2011 [Online]. Available online at: http://www.cdc.gov/foodnet/data/trends/tables/table~5.html (Accessed April 11 2013).
Centers for Disease Control and Prevention (CDC) (2015). Preliminary incidence and trends of infection with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. Sites, 2006–2014. Morb. Mortal. Week. Report 64, 495–499.
Cernicchiaro, N., Ives, S. E., Edrington, T. S., Nagaraja, T. G., and Renter, D. G. (2016). Efficacy of a Salmonella Siderophore receptor protein vaccine on fecal shedding and lymph node carriage of Salmonella in commercial feedlot cattle. Foodb. Pathogens Dis. 13, 517–525. doi: 10.1089/fpd.2016.2129
PubMed Abstract | CrossRef Full Text | Google Scholar
Clinical and Laboratory Standards Institute (2013). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Doerscher, D. R., Lutz, T. L., Whisenant, S. J., Smith, K. R., Morris, C. A., and Schroeder, C. M. (2015). Microbiological testing results of boneless and ground beef purchased for the national school lunch program, 2011 to 2014. J. Food Protect. 78, 1656–1663. doi: 10.4315/0362-028X.JFP-15-101
PubMed Abstract | CrossRef Full Text | Google Scholar
Edrington, T. S., Loneragan, G. H., Genovese, K. J., He, H., Callaway, T. R., Anderson, R. C., et al. (2013a). Development of a transdermal Salmonella challenge model in calves. J. Food Protect. 76, 1255–1258. doi: 10.4315/0362-028X.JFP-12-317
PubMed Abstract | CrossRef Full Text | Google Scholar
Edrington, T. S., Loneragan, G. H., Hill, J., Genovese, K. J., Brichta-Harhay, D. M., Farrow, R. L., et al. (2013b). Development of challenge models to evaulate the efficacy of a vaccine to reduce carriage of Salmonella in peripheral lymph nodes of cattle. J. Food Protect. 76, 1259–1263. doi: 10.4315/0362-028X.JFP-12-319
PubMed Abstract | CrossRef Full Text | Google Scholar
Food and Drug Administration (FDA) (2011). National Antimicrobial Resistance Monitoring System- Enteric Bacteria (NARMS): 2012 Retail Meat Report. Rockville, MD: U.S. Department of Health and Human Services, FDA.
Food Safety and Inspection Service (2011). NARMS Retail Meat Report, 2011. Washington, DC: U.S. Food and Drug Administration
Gragg, S. E., Loneragan, G. H., Brashears, M. M., Arthur, T. M., Bosilevac, J. M., Kalchayanand, N., et al. (2013a). Cross-sectional study examining Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at harvest. Foodb. Pathogens Dis. 10, 368–374. doi: 10.1089/fpd.2012.1275
PubMed Abstract | CrossRef Full Text | Google Scholar
Gragg, S. E., Loneragan, G. H., Nightingale, K. K., Brichta-Harhay, D. M., Ruiz, H., Elder, J. R., et al. (2013b). Substantial within-animal diversity of Salmonella isolates from lymph nodes, feces, and hides of cattle at slaughter. Appl. Environ. Microbiol. 79, 4744–4750. doi: 10.1128/AEM.01020-13
PubMed Abstract | CrossRef Full Text | Google Scholar
Grimont, P. A. D., and Weill, F.-X. (2007). Antigenic Formulae of the Salmonella Serovars, 9th Edn.. Paris: WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur. Available online at: http://www.pasteur.fr/sante/clre/cadrecnr/salmoms/WKLM_En.pdf
Guo, C. F., Hoekstra, R. M., Schroeder, C. M., Pires, S. M., Ong, K. L., Hartnett, E., et al. (2011). Application of bayesian techniques to model the burden of human salmonellosis attributable to U.S. food commodities at the point of processing: adaptation of a Danish model. Foodb. Pathog. Dis. 8, 509–516. doi: 10.1089/fpd.2010.0714
PubMed Abstract | CrossRef Full Text | Google Scholar
Haneklaus, A. N., Harris, K. B., Griffin, D. B., Edrington, T. S., Lucia, L. M., and Savell, J. W. (2012). Salmonella prevalence in bovine lymph nodes differs among feedyards. J. Food Protect. 75, 1131–1133. doi: 10.4315/0362-028X.JFP-11-530
PubMed Abstract | CrossRef Full Text | Google Scholar
Hanson, D., Loneragan, G., Brown, T., Nisbet, D., Hume, M., and Edrington, T. (2016). Evidence supporting vertical transmission of Salmonella in dairy cattle. Epidemiol. Infect. 144, 962–967. doi: 10.1017/S0950268815002241
PubMed Abstract | CrossRef Full Text | Google Scholar
Harvey, R. R., Friedman, C. R., Crim, S. M., Judd, M., Barrett, K. A., Tolar, B., et al. (2017). Epidemiology of Salmonella enterica Serotype Dublin Infections among Humans, United States, 1968–2013. Emerg. Infect. Dis. 23:1493. doi: 10.3201/eid2309.170136
PubMed Abstract | CrossRef Full Text | Google Scholar
Herrera-Leon, S., McQuiston, J. R., Usera, M. A., Fields, P. I., Garaizar, J., and Echeita, M. A. (2004). Multiplex PCR for distinguishing the most common phase-1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 42, 2581–2586. doi: 10.1128/JCM.42.6.2581-2586.2004
PubMed Abstract | CrossRef Full Text | Google Scholar
Jones, T. F., Ingram, L. A., Cieslak, P. R., Vugia, D. J., Tobin-D'Angelo, M., Hurd, S., et al. (2008). Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 198, 109–114. doi: 10.1086/588823
PubMed Abstract | CrossRef Full Text | Google Scholar
Koohmaraie, M., Scanga, J. A., De la Zerda, M. J., Koohmaraie, B., Tapay, L., Beskhlebnaya, V., et al. (2012). Tracking the sources of Salmonella in ground beef produced from Nonfed Cattle. J. Food Protect. 75, 1464–1468. doi: 10.4315/0362-028X.JFP-11-540
PubMed Abstract | CrossRef Full Text | Google Scholar
Kunze, D. J., Loneragan, G. H., Platt, T. M., Miller, M. F., Besser, T. E., Koohmaraie, M., et al. (2008). Salmonella enterica burden in harvest-ready cattle populations from the southern high plains of the United States. Appl. Environ. Microbiol. 74, 345–351. doi: 10.1128/AEM.02076-07
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, M., Malladi, S., Hurd, H. S., Goldsmith, T. J., Brichta-Harhay, D. M., and Loneragan, G. H. (2015). Salmonella spp. in lymph nodes of fed and cull cattle: relative assessment of risk to ground beef. Food Control 50, 423–434. doi: 10.1016/j.foodcont.2014.09.011
CrossRef Full Text | Google Scholar
McLaughlin, J. B., Castrodale, L. J., Gardner, M. J., Ahmed, R., and Gessner, B. D. (2006). Outbreak of multidrug-resistant Salmonella Typhimurium associated with ground beef served at a school potluck. J. Food Protect. 69, 666–670. doi: 10.4315/0362-028X-69.3.666
PubMed Abstract | CrossRef Full Text | Google Scholar
Morley, P. S., Dargatz, D. A., Hyatt, D. R., Dewell, G. A., Patterson, J. G., Burgess, B. A., et al. (2011). Effects of restricted antimicrobial exposure on antimicrobial resistance in fecal Escherichia coli from feedlot cattle. Foodb. Pathog. Dis. 8, 87–98. doi: 10.1089/fpd.2010.0632
PubMed Abstract | CrossRef Full Text | Google Scholar
Nucera, D. M., Maddox, C. W., Hoien-Dalen, P., and Weigel, R. M. (2006). Comparison of API 20E and invA PCR for identification of Salmonella enterica isolates from swine production units. J. Clin. Microbiol. 44, 3388–3390. doi: 10.1128/JCM.00972-06
PubMed Abstract | CrossRef Full Text | Google Scholar
Olafson, P. U., Brown, T. R., Lohmeyer, K. H., Harvey, R. B., Nisbet, D. J., Loneragan, G. H., et al. (2016). Assessing transmission of Salmonella to bovine peripheral lymph nodes upon horn fly feeding. J. Food Protect. 79, 1135–1142. doi: 10.4315/0362-028X.JFP-15-414
PubMed Abstract | CrossRef Full Text | Google Scholar
Rahn, K., Degrandis, S. A., Clarke, R. C., McEwen, S. A., Galan, J. E., Ginocchio, C., et al. (1992). Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Prob. 6, 271–279. doi: 10.1016/0890-8508(92)90002-F
PubMed Abstract | CrossRef Full Text | Google Scholar
Rao, S., Van Donkersgoed, J., Bohaychuk, V., Besser, T., Song, X.-M., Wagner, B., et al. (2010). Antimicrobial drug use and antimicrobial resistance in enteric bacteria among cattle from Alberta feedlots. Foodb. Pathog. Dis. 7, 449–457. doi: 10.1089/fpd.2009.0400
PubMed Abstract | CrossRef Full Text | Google Scholar
Rodriguez-Rivera, L. D., Cummings, K. J., Loneragan, G. H., Rankin, S. C., Hanson, D. L., Leone, W. M., et al. (2016). Salmonella prevalence and antimicrobial susceptibility among dairy farm environmental samples collected in Texas. Foodb. Pathog. Dis. 13, 205–211. doi: 10.1089/fpd.2015.2037
PubMed Abstract | CrossRef Full Text | Google Scholar
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
PubMed Abstract | CrossRef Full Text | Google Scholar
Schneider, J., White, P., Weiss, J., Norton, D., Lidgard, J., Gould, L., et al. (2011). Multistate outbreak of multidrug-resistant Salmonella Newport infections associated with ground beef, October to December 2007. J. Food Protect. 74, 1315–1319. doi: 10.4315/0362-028X.JFP-11-046
PubMed Abstract | CrossRef Full Text | Google Scholar
Sjölund-Karlsson, M., Joyce, K., Blickenstaff, K., Ball, T., Haro, J., Medalla, F. M., et al. (2011). Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrob. Agents Chemother. 55, 3985–3989. doi: 10.1128/AAC.00590-11
PubMed Abstract | CrossRef Full Text | Google Scholar
Sofos, J. N., Kochevar, S. L., Bellinger, G. R., Buege, D. R., Hancock, D. D., Ingham, S. C., et al. (1999). Sources and extent of microbiological contamination of beef carcasses in seven United States slaughtering plants. J. Food Protect. 62, 140–145. doi: 10.4315/0362-028X-62.2.140
PubMed Abstract | CrossRef Full Text | Google Scholar
Sorensen, O., Van Donkersgoed, J., McFall, M., Manninen, K., Gensler, G., and Ollis, G. (2002). Salmonella spp. shedding by Alberta beef cattle and the detection of Salmonella spp. in ground beef. J. Food Protect. 65, 484–491. doi: 10.4315/0362-028X-65.3.484
PubMed Abstract | CrossRef Full Text | Google Scholar
Suez, J., Porwollik, S., Dagan, A., Marzel, A., Schorr, Y. I., Desai, P. T., et al. (2013). Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS ONE 8:e58449. doi: 10.1371/journal.pone.0058449
PubMed Abstract | CrossRef Full Text | Google Scholar
Wells, S., Fedorka-Cray, P., Dargatz, D., Ferris, K., and Green, A. (2001). Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. J. Food Protect. 64, 3–11. doi: 10.4315/0362-028X-64.1.3
PubMed Abstract | CrossRef Full Text | Google Scholar
Wheeler, T. L., Kalchayanand, N., and Bosilevac, J. M. (2014). Pre- and post-harvest interventions to reduce pathogen contamination in the U.S. beef industry. Meat Sci. 98, 372–382. doi: 10.1016/j.meatsci.2014.06.026
PubMed Abstract | CrossRef Full Text | Google Scholar
Source: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02214/full
0 Response to "Beef Cattle Lymph Nodes Texas am University Prevalence"
Post a Comment